Creating Molecules from Smiles Strings#
Smiles strings provide a convenient way to represent a molecule as text.
You can create a molecule from a smile string using the
sire.smiles() function.
>>> import sire as sr
>>> mol = sr.smiles("C1:C:C:C:C:C1")
>>> print(mol.atoms())
Selector<SireMol::Atom>( size=12
0: Atom( C1:1 [ -0.92, -1.05, 0.02] )
1: Atom( C2:2 [ -1.37, 0.27, -0.04] )
2: Atom( C3:3 [ -0.45, 1.32, -0.06] )
3: Atom( C4:4 [ 0.92, 1.05, -0.02] )
4: Atom( C5:5 [ 1.37, -0.27, 0.04] )
...
7: Atom( H8:8 [ -2.43, 0.48, -0.07] )
8: Atom( H9:9 [ -0.80, 2.35, -0.11] )
9: Atom( H10:10 [ 1.63, 1.87, -0.04] )
10: Atom( H11:11 [ 2.43, -0.48, 0.07] )
11: Atom( H12:12 [ 0.80, -2.35, 0.11] )
)
>>> mol.view()
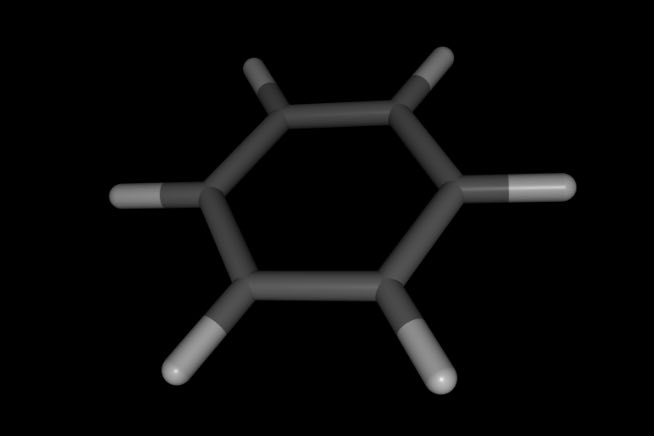
Note how hydrogen atoms and coordinates of all atoms have
been generated automatically. You can control this using
the add_hydrogens and generate_coordinates options, e.g.
>>> mol = sr.smiles("C1:C:C:C:C:C1", add_hydrogens=False,
... generate_coordinates=False)
>>> print(mol.atoms())
Selector<SireMol::Atom>( size=6
0: Atom( C1:1 )
1: Atom( C2:2 )
2: Atom( C3:3 )
3: Atom( C4:4 )
4: Atom( C5:5 )
5: Atom( C6:6 )
)
Note
Note that hydrogens will automatically be added if
generate_coordinates is True (which is the default)