Moving Bonds, Angles and Dihedrals¶
Moving internals (e.g. bonds, angles and dihedrals) within molecules is also straightforward. You do this via a cursor that edits the internal (or internals).
Moving Bonds¶
For example, here we will get a cursor that edits all of the carbon-hydrogen bonds in the first molecule.
>>> mols = sr.load(sr.expand(sr.tutorial_url, ["ala.top", "ala.crd"]))
>>> cursor = mols[0].bonds("element C", "element H").cursor()
Cursors that edit bonds have two additional key functions;
set_length/set_lengthsandchange_length/change_lengths
These functions either set the bond length to a specified value, or change the bond length by the specified delta.
For example, we can set the lengths of all of the carbon-hydrogen
bonds to 1 Å using set_length
>>> cursor.set_length(1.0)
>>> print(cursor.lengths())
[1 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å]
Note
Note that you can specify units of length if you want, e.g.
set_length(1 * sr.units.angstrom). The default length units
are used if units aren’t specified.
Note
Note also that set_length sets the length of all bonds edited
by the cursor to the passed length. Use set_lengths and pass
in a list of lengths if you want to set the bonds to
different lengths.
We can change the length of the third carbon-hydrogen bond by 0.5 Å using
change_length, e.g.
>>> cursor[2].change_length(0.5)
>>> print(cursor[2].length())
1.5 Å
>>> print(cursor.lengths())
[1 Å, 1 Å, 1.5 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å, 1 Å]
Tracking movement with trajectories¶
You can visualise and analyse the movements you are performing by saving trajectory frames. For example, lets gradually stretch the carbon-carbon bond in the first molecule, saving trajectory frames as we go.
>>> mol = mols[0]
>>> cursor = mol.cursor()
>>> cursor.save_frame()
>>> bond_cursor = cursor.bond("atomname CA", "resname ALA and atomname C")
>>> for i in range(0, 10):
... bond_cursor.change_length(0.05)
... bond_cursor.save_frame()
>>> mol = cursor.commit()
You can view a movie of this movement using
>>> mol.view()
You can get the energy for each frame of the trajectory using
>>> mol.trajectory().energy()
frame time 1-4_LJ 1-4_coulomb angle bond dihedral \
0 0 0.0 2.920343 44.880519 6.791894 4.548210 9.532594
1 1 0.0 2.607594 43.652433 6.791894 6.645588 9.532594
2 2 0.0 2.366888 42.448164 6.791894 10.327967 9.532594
3 3 0.0 2.182746 41.267230 6.791894 15.595345 9.532594
4 4 0.0 2.043079 40.109161 6.791894 22.447723 9.532594
5 5 0.0 1.938407 38.973497 6.791894 30.885102 9.532594
6 6 0.0 1.861278 37.859784 6.791894 40.907480 9.532594
7 7 0.0 1.805813 36.767577 6.791894 52.514858 9.532594
8 8 0.0 1.767366 35.696436 6.791894 65.707237 9.532594
9 9 0.0 1.742254 34.645928 6.791894 80.484615 9.532594
10 10 0.0 1.727556 33.615627 6.791894 96.846993 9.532594
improper intra_LJ intra_coulomb total
0 0.285078 -1.102960 -46.370186 21.485492
1 0.285078 -1.076241 -45.394975 23.043966
2 0.285078 -1.046408 -44.442886 26.263291
3 0.285078 -1.014466 -43.513222 31.127199
4 0.285078 -0.981224 -42.605298 37.623007
5 0.285078 -0.947316 -41.718473 45.740783
6 0.285078 -0.913235 -40.852145 55.472728
7 0.285078 -0.879364 -40.005697 66.812753
8 0.285078 -0.845992 -39.178556 79.756056
9 0.285078 -0.813341 -38.370160 94.298863
10 0.285078 -0.781567 -37.579977 110.438198
Moving Angles¶
We can move angles in our molecule in a similar way. In this case,
we use the set_size and change_size functions of the
Cursor.
>>> mol = mols[0]
>>> cursor = mol.cursor().angles("element C", "element N", "element C")
>>> print(cursor.sizes())
[123.725°, 123.802°]
>>> cursor.set_size(120)
>>> print(cursor.sizes())
[120°, 120°]
Note
You can specify the angle units, e.g. cursor.set_size(125*sr.units.degrees).
If you don’t specify the unit, then the default unit (degrees) is used.
The set_sizes and change_sizes functions enable you to set the
angles to different sizes in a single function call, e.g.
>>> cursor.set_sizes([115, 125])
>>> print(cursor.sizes())
[115°, 125°]
You could visualise the movement in the same way as you did when moving bonds.
>>> mol = mols[0]
>>> cursor.set_size(100)
>>> cursor.save_frame()
>>> for i in range(0, 10):
... cursor.change_size(4)
... cursor.save_frame()
>>> mol = cursor.commit()
>>> mol.view()
You could get the energy for each frame using
>>> mol.trajectory().energy()
frame time 1-4_LJ 1-4_coulomb angle bond dihedral \
0 0 0.0 30.900211 51.456752 37.326211 4.54821 9.533890
1 1 0.0 18.891062 50.151760 27.403285 4.54821 9.533543
2 2 0.0 11.953834 48.940301 19.412892 4.54821 9.533266
3 3 0.0 7.847938 47.811189 13.358399 4.54821 9.533042
4 4 0.0 5.364667 46.754576 9.242185 4.54821 9.532859
5 5 0.0 3.834074 45.761769 7.065959 4.54821 9.532710
6 6 0.0 2.875548 44.825062 6.830962 4.54821 9.532587
7 7 0.0 2.268087 43.937622 8.538100 4.54821 9.532486
8 8 0.0 1.881119 43.093390 12.188025 4.54821 9.532405
9 9 0.0 1.636824 42.287022 17.781193 4.54821 9.532341
10 10 0.0 1.489374 41.513861 25.317903 4.54821 9.532292
improper intra_LJ intra_coulomb total
0 0.317684 61.043125 -55.446533 139.679549
1 0.308098 24.833982 -53.481881 82.188059
2 0.300869 10.163214 -51.731143 53.121443
3 0.295320 3.804292 -50.161684 37.036707
4 0.291006 0.901951 -48.747455 27.888000
5 0.287623 -0.470122 -47.467360 23.092862
6 0.284961 -1.130151 -46.304294 21.462885
7 0.282872 -1.446705 -45.244352 22.416321
8 0.281249 -1.594252 -44.276305 25.653841
9 0.280017 -1.658877 -43.391193 31.015537
10 0.279122 -1.684468 -42.582148 38.414145
Moving Dihedrals¶
Dihedrals are moved using a similar syntax as bonds and angles. You use
set_size / set_sizes to set dihedrals, and
change_size / change_sizes to change dihedrals. For example;
>>> mol = mols[0]
>>> cursor = mol.cursor().dihedrals("element N", "element C",
"element C", "element N")
>>> print(cursor.sizes())
[163.039°]
>>> cursor.set_size(150)
>>> print(cursor.sizes())
[150°]
You can view a movie of the changes by saving frames to a trajectory, e.g.
>>> mol = mols[0]
>>> cursor = mol.cursor().dihedrals("element N", "element C",
"element C", "element N")
>>> cursor.save_frame()
>>> for i in range(0, 12):
... cursor.change_size(30)
... cursor.save_frame()
>>> mol = cursor.commit()
>>> mol.view()
You can get the energies for each frame using;
>>> print(mol.trajectory().energy())
frame time 1-4_LJ 1-4_coulomb angle bond dihedral \
0 0 0.0 2.920343 44.880519 6.791894 4.54821 9.532594
1 1 0.0 2.903734 45.413449 6.791894 4.54821 10.206761
2 2 0.0 2.866713 46.060627 6.791894 4.54821 10.253447
3 3 0.0 2.879910 47.150792 6.791894 4.54821 9.674494
4 4 0.0 3.215898 48.524346 6.791894 4.54821 10.327498
5 5 0.0 3.605072 49.294830 6.791894 4.54821 10.980599
6 6 0.0 3.431110 48.620810 6.791894 4.54821 9.843168
7 7 0.0 3.221010 46.739679 6.791894 4.54821 9.406235
8 8 0.0 3.778984 44.691981 6.791894 4.54821 12.445042
9 9 0.0 4.696463 43.463628 6.791894 4.54821 15.872254
10 10 0.0 4.364056 43.437424 6.791894 4.54821 14.982017
11 11 0.0 3.347161 44.154732 6.791894 4.54821 11.243423
12 12 0.0 2.920343 44.880519 6.791894 4.54821 9.532594
improper intra_LJ intra_coulomb total
0 0.285078 -1.102960 -46.370186 21.485492
1 0.285078 -1.054636 -46.361446 22.733043
2 0.285078 -1.236196 -46.037819 23.531954
3 0.285078 -1.485350 -45.416578 24.428450
4 0.285078 7.543340 -42.220497 39.015767
5 0.285078 321.043305 -32.911014 363.637974
6 0.285078 246.990282 -31.962216 288.548335
7 0.285078 3.895676 -38.766103 36.121679
8 0.285078 6.624047 -40.837995 38.327241
9 0.285078 19.768052 -41.352134 54.073444
10 0.285078 54.297987 -43.297191 85.409475
11 0.285078 10.276564 -45.440864 35.206198
12 0.285078 -1.102964 -46.370172 21.485502
Moving individual dihedrals¶
You may have noticed that all of the atoms around a dihedral were rotated when you changed an individual dihedral. This is because, by default, the move rotates the bond that lies at the center of the dihedral.
You can rotate only the specified dihedral by setting the move_all
option to False. For example;
>>> mol = mols[0]
>>> cursor = mol.cursor().dihedrals("element N", "element C",
"element C", "element N")
>>> cursor.save_frame()
>>> for i in range(0, 12):
... cursor.change_size(30, move_all=False)
... cursor.save_frame()
>>> mol = cursor.commit()
>>> mol.view()
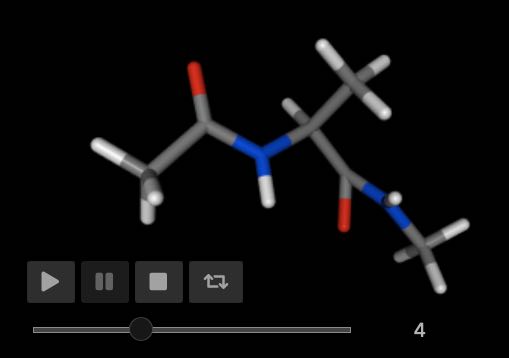
Notice how this results in a clash between the nitrogen (which is being rotated) and the oxygen (which is not rotated).
In general, to avoid atom clashes, it is normally better to rotate all
atoms about the central dihedral bond. As such, move_all defaults
to True.
Aligning, Anchoring and Weighting¶
There are a few more options that can be used to give more fine-grained control over how the molecule is moved.
The first is auto_align. This defaults to True, and switches
on automatic re-alignment of the molecule against itself after the
move. This is a simple alignment that minimises the root mean square
deviation (RMSD) between the molecule before and after the move.
Set auto_align to False if you want to switch off this
automatic alignment.
The next option is weighting. The internal moves work by dividing
the molecule into two sets about the bond (or bonds) involved in the move.
For example, to rotate the N-C1-C2-N dihedral as we did above, the molecule
is split into two sets about the central C1-C2 bond. The “left” set
contains all atoms that are bonded to C1, or which can trace their
connectivity to C1 through other atoms. The “right” set contains
all atoms that are bonded to C2, or which can trace their connectivity
to C2 through other atoms.
The rotation is achieved by rotating the “left” and “right” sets by opposite
amounts around the vector of the C1-C2 bond, depending on the “weight”
of the two sets. For example, if the dihedral was to be rotated by 30°,
and both sets were weighted equally, then one set would be rotated
by +15°, whle the other would be rotated by -15°.
The weighting option sets the algorithm that is used to weight the
rotation (or translation in the case for bond moves). Valid options are;
relative_mass: the change is weighted linearly according to the relative mass of the two sets. The lightest set will be moved the most, while the heaviest set would move the least.absolute_mass: the change will entirely be weighted towards the set with the lightest mass. The lightest set will receive all of the move, while the heaviest set will not be moved.relative_number: the change is weighted linearly according to the relative number of atoms in both sets. The set with fewest atoms will be moved the most, while the set with most atoms will move the least.absolute_number: the change will entirely be weighted towards the set with the fewest atoms. The set with the fewest atoms will receive all of the move, while the set with the most atoms will not be moved.
Finally, you can specify anchor atoms using the anchor option.
This specifies atoms which are anchored, and which should not be moved
at all when you stretch a bond, or change an angle or dihedral. The
anchor option accepts any value that would be supported by
the indexing operator of the view being moved. This will be used to index the
view being moved, to get the set of atoms that must remain immobile.
If one of the sets of atoms to be moved contains an anchor, then
it will not be moved, and the full weight of the move will be applied
to the other set. Note that it is an error to have anchors in both sets,
as this would prevent the move from being made. An exception will be
raised in this case. Note also that setting the anchor option will
automatically disable automatic alignment.
For example;
>>> cursor.change_size(30, move_all=True,
weighting="relative_mass",
auto_align=False)
would rotate all of the atoms around a dihedral by 30°, distributing
the move according to the relative_mass algorithm. The molecule would
not be automatically re-aligned after the move.
or;
>>> cursor.bonds("element C", "element H").set_length(1, anchor="element C")
would set all carbon-hydrogen bonds to a length of 1 Å, while placing
anchors on all the carbon atoms. This ensures that only the hydrogens
will be moved. The use of anchors means that auto_align is set to
False, and so the molecule would not be automatically re-aligned
after the move.